Normal form demo - bifurcation detection and normal form computation
This file is self-contained and requires the extension ddebiftool_extra_nmfm.
$Id$
Contents
The system
This system represents a non-dimensionalized model of two interacting layers of neurons.
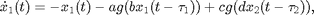
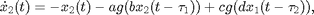
where 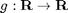 is of the sigmoidal form
is of the sigmoidal form
![$$ g(z) = \left[\tanh(z-1) + \tanh(1)\right]\cosh(1)^2. $$](nmfm_demo_eq61256.png)
The variables 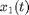 and
and 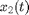 represent the population-averaged neural activity at time
represent the population-averaged neural activity at time  in layers one and two, respectively. The parameter
in layers one and two, respectively. The parameter 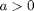 is a measure of the strength of inhibitory feedback, while
is a measure of the strength of inhibitory feedback, while 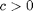 measures the strength of the excitatory effect of one layer on the other. The parameters
measures the strength of the excitatory effect of one layer on the other. The parameters 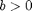 and
and 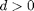 are saturation rates and the delays
are saturation rates and the delays 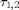 represent time lags in the inhibitory feedback loop and excitatory inter-layer connection. Note that the system is symmetric with respect to interchanging the labels
represent time lags in the inhibitory feedback loop and excitatory inter-layer connection. Note that the system is symmetric with respect to interchanging the labels  and
and  , so equilibria are necessarily of the form
, so equilibria are necessarily of the form 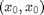 .
.
Initialization
First, we load the necessary DDE-Biftool extensions. We use existing files in the funcs structure. These files were generated using a Maple script. Lastly, we define the boundaries and step sizes of the active parameters.
clear variables; close all; addpath('../../ddebiftool',... '../../ddebiftool_extra_psol',... '../../ddebiftool_extra_nmfm',... '../../ddebiftool_utilities'); funcs=set_funcs(... 'sys_rhs', @(xx,par) sys_rhs(xx,par),... 'sys_tau', @() sys_tau(),... 'sys_deri', @(xx,par,nx,np,v) sys_deri(xx,par,nx,np,v),... 'sys_mfderi',@(xx,par,varargin) sys_mfderi(xx,par,varargin{:})); n = 2; astepsize = 0.0005; cstepsize = 0.005; amin = 0.0; amax = 0.55; cmin = 0.0; cmax = 1.0;
Continuation and plot of steady state branch
We set up a steady state branch using SetupStst and do a standard continuation. Then, we plot the branch.
branch1 = SetupStst(funcs,'x',[0;0],'parameter',[0.25, 2, 15/29, 1.2, 12.7, 20.2],... 'contpar',1,'max_step',[1 astepsize],'max_bound',[1 amax],'min_bound',[1 amin],... 'newheuristics_tests',0); branch1.method.continuation.plot = 0; [branch1,s,f,r] = br_contn(funcs,branch1,300); branch1 = br_rvers(branch1); [branch1,s,f,r] = br_contn(funcs,branch1,300); figure; [xm,ym] = df_measr(0,branch1,1); br_plot(branch1,xm,ym); xlabel('a'); ylabel('x_1');
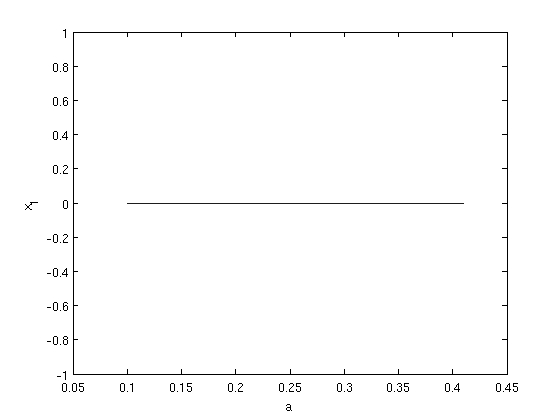
Bifurcation detection on the steady state branch
To detect bifurcations, we have to call the function br_bifdet.
function [newbranch, success] = br_bifdet(funcs, branch)
Inputs:
- funcs: problem-definition structure with user-defined functions
- branch: branch of steady state (stst) points
Outputs:
- newbranch: branch with flagged bifurcation points containing normal form coefficient structures
- success: flag whether detection was successful
To adjust the behavior of this algorithm, you can change a number of parameters in branch.method.bifurcation:
- minimal_real_part: only roots with a real part above this parameter will be taken into consideration when detecting bifurcations. The default is -0.03. Too many roots will cause overflow (because the hopf detector multiplies them all), so if an overflow is reported, try increasing this parameter.
- correction_tolerance: to correct hopf bifurcations, the standard routine p_correc is used. The default value for this corrector is 1e-7.
- secant_iterations: codimension 2 bifurcations are corrected using a secant method. This is the maximum number of iterations, with default value 30.
- secant_tolerance: this is the tolerance of the secant method, with default 1e-9.
- radial_tolerance_factor: after a bifurcation point has been corrected, it is checked whether the corrected point actually lies on the branch. If the bifurcation point originated from a change in the roots between points A and B, the new point is considered "on the branch" if its distance to the line AB is less than 25% of length(AB). This default percentage can be adjusted with this parameter.
You will see some notifications about found hopf points not falling within the branch. This indicates a false positive: the correction algorithm produced a hopf point, but it ended up some distance away from the branch.
branch1.method.bifurcation.minimal_real_part = -0.03;
branch1.method.bifurcation.secant_tolerance=1e-5;
branch1.method.bifurcation.correction_tolerance=1e-5;
fprintf('Bifurcation detection\n');
branch1 = br_bifdet(funcs, branch1);
Bifurcation detection BR_BIFDET: hopf point found at par(1) = 0.3676281680. BR_BIFDET: L1 = -0.0028635604, omega = 0.7234938546, par(1) = 0.3676281680. BR_BIFDET: hopf point found at par(1) = 0.3453112603. BR_BIFDET: L1 = -0.0133009607, omega = 0.1826428346, par(1) = 0.3453112603. BR_BIFDET: hopf point found at par(1) = 0.2988605971. BR_BIFDET: L1 = -0.0054576675, omega = 0.2710841446, par(1) = 0.2988605971. BR_BIFDET: hopf point found at par(1) = 0.2988605971. BR_BIFDET: the detected hopf point does not fall within the branch. BR_BIFDET: hopf point found at par(1) = 0.2988605971. BR_BIFDET: the detected hopf point does not fall within the branch.
Extracting bifurcation locations
As you can see, three Hopf points were detected. They will have been flagged, e.g. branch1.point(132).flag = 'hopf'. They will also have normal form information: branch1.point(132).nmfm.L1 = -0.0133009607. We will choose two hopf points and continue them. (These two were chosen because their branches happen to intersect, producing a Double Hopf bifurcation.)
To this end, we need to know where the bifurcations were found. We do this by calling br_getflags on the steady state branch.
flagged_point_indices = br_getflags(branch)
Inputs:
- branch: any branch
Outputs:
- flagged_point_indices: a matrix containing the indices of the bifurcation points in branch.
The way the FPI (flagged point indices) matrix is structured is as follows. Every bifurcation has its own number (0 = stst, 1 = hopf, ...). You can convert between these representations by using string = num2bif(number) and number = bif2num(string). Now, FPI(bif2num('bifucation type'),:) is a list of the indices at which the bifurcation of the designated type occurs. (For the full list, you can refer to any of these two files.)
We first select the second Hopf point on the steady state branch.
FPI = br_getflags(branch1);
start_ind = FPI(bif2num('hopf'),2);
Continuation of the first Hopf branch
We do a standard continuation, using the starting index obtained from the flagged point indices.
fprintf('----- Hopf branch 1 -----\n'); [branch2, suc] = SetupHopf(funcs, branch1, start_ind, 'contpar', [1 3], 'dir', 3, 'step', cstepsize); branch2.parameter.min_bound=[5 0; 6 0; 3 cmin; 1 amin]; branch2.parameter.max_bound=[1 amax; 3 cmax]; branch2.parameter.max_step=[1 0.002; 3 cstepsize]; branch2.method.continuation.plot = 0; branch2.method.stability.minimal_time_step = 0.005; % default 0.01 [branch2,s,f,r]=br_contn(funcs,branch2,300); branch2 = br_rvers(branch2); [branch2,s,f,r]=br_contn(funcs,branch2,300);
----- Hopf branch 1 ----- BR_CONTN warning: boundary hit. BR_CONTN warning: boundary hit.
Bifurcation detection on the first Hopf branch
We repeat the steps for bifurcation detection, but now apply it to the hopf branch. This yields several higher-order bifurcations.
branch2.method.bifurcation.minimal_real_part = -0.03; branch2.method.bifurcation.secant_tolerance=1e-5; branch2.method.bifurcation.correction_tolerance=1e-5; branch2 = br_bifdet(funcs,branch2);
BR_BIFDET: zeho point found at par(1) = 0.0037927481, par(3) = 0.8396720010. BR_BIFDET: omega = 0.1485568996, par(1) = 0.0037927481, par(3) = 0.8396720010. s = 0.0000855602, theta = 2.0242706451. BR_BIFDET: hoho point found at par(1) = 0.0899922748, par(3) = 0.7735178974. BR_BIFDET: omega1 = 0.1560058562, omega2 = 0.2900084424, par(1) = 0.0899922748, par(3) = 0.7735178974. theta(0) = 1.5255011842, delta(0) = 1.6984697752. BR_BIFDET: genh point found at par(1) = 0.2566137217, par(3) = 0.6210725697. BR_BIFDET: L2 = 0.0018108090, omega = 0.1722171486, par(1) = 0.2566137217, par(3) = 0.6210725697. BR_BIFDET: Unable to correct as hoho point near par(1) = 0.2813. BR_BIFDET: Unable to correct as zeho point near par(1) = 0.3652. BR_BIFDET: Unable to correct as hoho point near par(1) = 0.4966.
The second Hopf branch
We now repeat the entire exercise, but with the third hopf point as input.
fprintf('----- Hopf branch 2 -----\n'); FPI = br_getflags(branch1); start_ind = FPI(bif2num('hopf'),3); [branch3, suc] = SetupHopf(funcs, branch1, start_ind, 'contpar', [1 3], 'dir', 3, 'step', cstepsize); branch3.parameter.min_bound=[5 0; 6 0; 3 cmin; 1 amin]; branch3.parameter.max_bound=[1 amax; 3 cmax]; branch3.parameter.max_step=[1 0.002; 3 cstepsize]; branch3.method.continuation.plot = 0; branch3.method.stability.minimal_time_step = 0.005; % default 0.01 [branch3,s,f,r]=br_contn(funcs,branch3,300); branch3 = br_rvers(branch3); [branch3,s,f,r]=br_contn(funcs,branch3,300); branch3.method.bifurcation.minimal_real_part = -0.03; branch3.method.bifurcation.secant_tolerance=1e-5; branch3.method.bifurcation.correction_tolerance=1e-5; branch3 = br_bifdet(funcs, branch3);
----- Hopf branch 2 ----- BR_CONTN warning: boundary hit. BR_CONTN warning: boundary hit. BR_BIFDET: zeho point found at par(1) = 0.0133327150, par(3) = 0.8554382654. BR_BIFDET: omega = 0.2958015628, par(1) = 0.0133327150, par(3) = 0.8554382654. s = 0.0000803411, theta = 2.0969328059. BR_BIFDET: hoho point found at par(1) = 0.0900149247, par(3) = 0.7736041399. BR_BIFDET: omega1 = 0.2900073517, omega2 = 0.1560072608, par(1) = 0.0900149247, par(3) = 0.7736041399. theta(0) = 1.6984018466, delta(0) = 1.5254221873. BR_BIFDET: genh point found at par(1) = 0.2518504300, par(3) = 0.5812203535. BR_BIFDET: L2 = 0.0011029849, omega = 0.2759107995, par(1) = 0.2518504300, par(3) = 0.5812203535. BR_BIFDET: Unable to correct as zeho point near par(1) = 0.3348.
Plotting the bifurcations on the branches
We now plot the branches. On the hopf branches, we want to plot the bifurcation points with a special marker. This is done using the function br_bifplot.
function br_bifplot(branch,x_m,y_m,method)
Inputs:
- branch: any branch (containing flagged bifurcation points)
- x_m: measure for the x-axis
- y_m: measure for the y-axis
- method: a structure specifying a marker type for all bifurcation types
A default bifurcation plot structure can be obtained with df_bifplot. This function produces a structure that plots all bifurcations with 'o' and steady state points with '-'.
figure; bifplot = df_bifplot(); bifplot.hoho = '*'; [xm,ym] = df_measr(0,branch2,1); br_plot(branch2,xm,ym,'-'); br_bifplot(branch2,xm,ym,bifplot); [xm,ym] = df_measr(0,branch3,1); br_plot(branch3,xm,ym,'-'); br_bifplot(branch3,xm,ym,bifplot); br_plot(branch1,xm,ym,'--'); xlabel('a'); ylabel('c'); return;
